OUR OFFERINGS
Case Study
Small Molecule Design For Neuronal Disorder
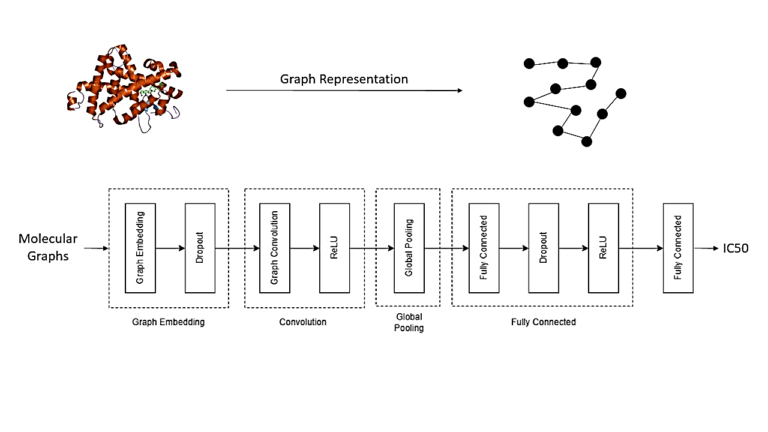
The client approached us with the objective to optimize their drug discovery process, aiming to reduce costs and accelerate the timeline for bringing new drugs to the advanced phase. Our team proposed the integration of in silico AI-assisted drug design solution to complement experimental methods and enhance the overall drug development pipeline.

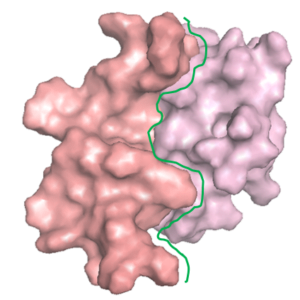
Peptide Design to Inhibit Protein Aggregation
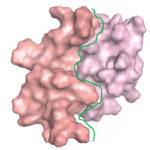
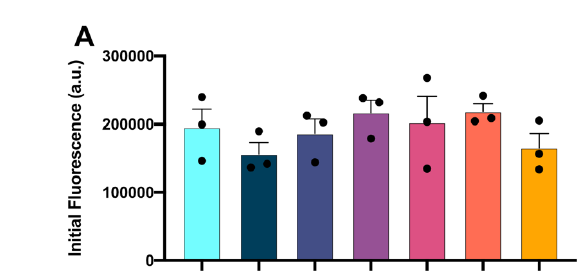
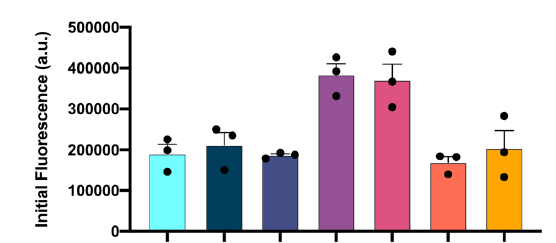
A protein ‘X’ forms an aggregate and the deposition of these aggregates results in a disorder. client Biotech was looking for a peptide/oregano peptide that can bind at the interface of the oligomer/dimer and inhibit the initiation of the aggregation process.
Identifying & Targeting new Target for Infectious Diseases
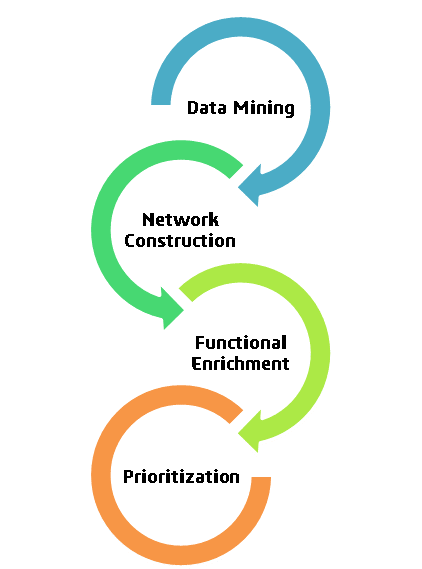
Client’s objective was expediting drug development and requiring drug targets for infectious disease ‘A’, so we proposed a network-based drug target identification approach. Leveraging in silico techniques and approved drugs, our innovative strategy aims to significantly reduce the time and cost associated with the process. This approach holds promise for rapidly identifying potential treatments, accelerating the quest for effective therapies against disease ‘A’.